topiary
Python framework for doing ancestral sequence reconstruction
Ancestral sequence reconstruction (ASR) is a powerful method to study protein evolution. It requires constructing a multiple sequence alignment, running a software pipeline with several software packages, and converting between arcane file types. Topiary streamlines this process, simplifying the workflow and helping non-experts do best-practice ASR.
Features
Automatic. Performs sequence database construction, quality control, multiple sequence alignment, tree construction, gene/species tree reconciliation, and ancestral reconstruction with minimal user input.
Human-oriented. Users prepare their input as spreadsheets, not complicated text files. Outputs are spreadsheets and graphical summaries of ancestor quality.
Species aware. Integrates with the Open Tree of Life database, improving selection of sequences and tree/ancestor inference.
Flexible. Use as a command line program or do custom analyses and plotting using the topiary API in a Jupyter notebook or Python script.
Modern. Topiary is built around a collection of modern, actively-supported, phylogenetic software tools: OpenTree, muscle 5, RAxML-NG, GeneRax, PastML, and toytree.
Steps done by topiary
Try it out on Google Colab
Workflow
Topiary automates the computational steps of an ASR calculation, allowing the user to focus on the three steps that require human insight: defining the problem, validating the alignment, and characterizing the resulting ancestors. The graphic below shows the steps done by the user (brain icons) versus software (tree icons) in a topiary calculation.
Example input/output
User input to a topiary calculation
A user prepares a “seed dataframe” setting the scope for the calculation.
name |
aliases |
species |
sequence |
LY96 |
ESOP1;Myeloid Differentiation Protein-2;MD-2;lymphocyte antigen 96;LY-96 |
Homo sapiens |
MLPFLFF… |
LY96 |
ESOP1;Myeloid Differentiation Protein-2;MD-2;lymphocyte antigen 96;LY-96 |
Danio rerio |
MALWCPS… |
LY86 |
Lymphocyte Antigen 86;LY86;Myeloid Differentiation Protein-1;MD-1;RP105-associated 3;MMD-1 |
Homo sapiens |
MKGFTAT… |
LY86 |
Lymphocyte Antigen 86;LY86;Myeloid Differentiation Protein-1;MD-1;RP105-associated 3;MMD-1 |
Danio rerio |
MKTYFNM… |
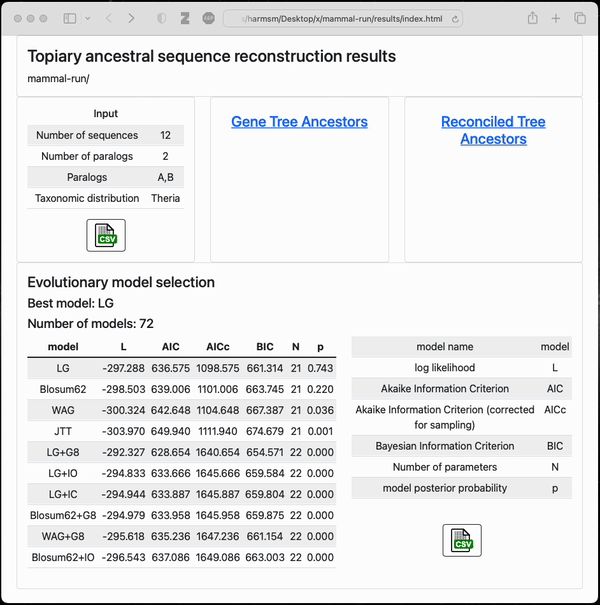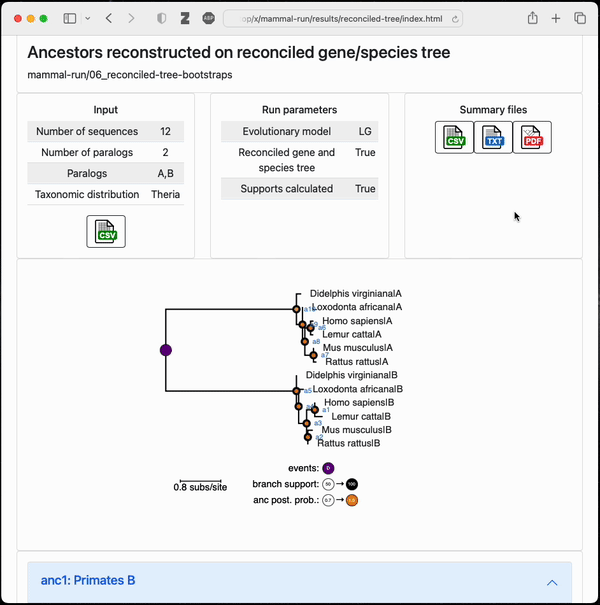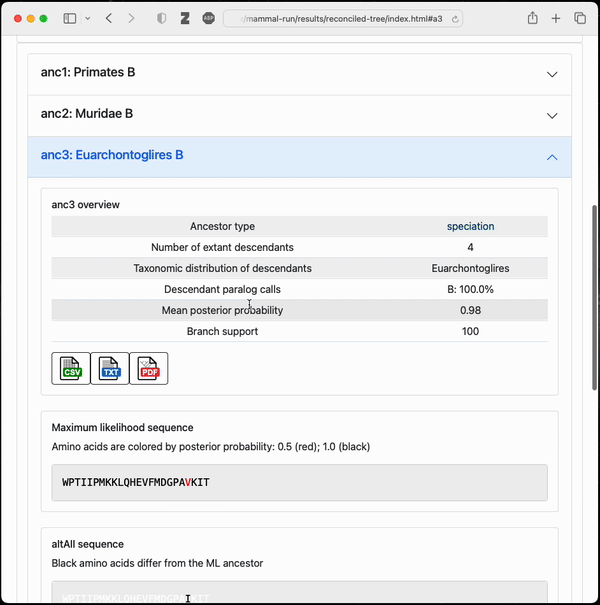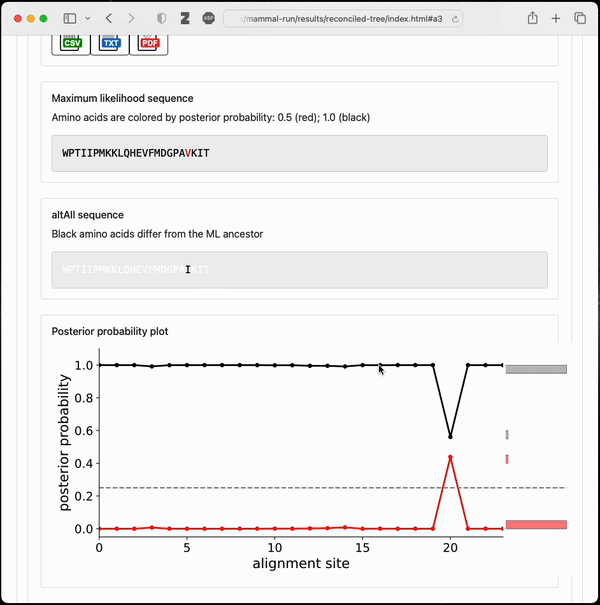
Final output from a small example topiary calculation
After running the pipeline, topiary returns a shareable directory with an html summary of all results.
Installation
See the installation page.
Short protocol
For a more detailed protocol, see the protocol page.
Create a seed spreadsheet with a handful of sequences that define the scope of the ASR study. For examples, see the table above or download the full example.
Construct a multiple sequence alignment from a the seed spreadsheet (“seed.xlsx”, for example). This can be run on a local computer or a cluster.
topiary-seed-to-alignment seed.xlsx --out_dir outputIf desired, visually inspect and edit the alignment in an external alignment viewer. (We recommend aliview.) Load the edited alignment into a topiary dataframe.
topiary-load-fasta-into output/dataframe.csv edited_fasta final-dataframe.csvBuild a species-reconciled phylogenetic tree and infer ancestral sequences. This is usually run on a cluster.
topiary-alignment-to-ancestors final-dataframe.csv --out_dir ali_to_ancGenerate bootstrap replicates to measure branch supports. This is usually run on a cluster.
topiary-bootstrap-reconcile ali_to_anc num_threads
How to cite
If you use topiary in your research, please cite:
Orlandi KN*, Phillips SR*, Sailer ZR, Harman JL, Harms MJ. “Topiary: pruning the manual labor from ancestral sequence reconstruction” (2022) Protein Science 10.1002/pro.4551.
* Authors contributed equally
Please make sure to cite the tools we use in the package as well:
Muscle: Edgar RC (2021) bioRxiv https://doi.org/10.1101/2021.06.20.449169.
RAxML-NG: Kozlov et al (2019) Bioinformatics 35(21):4453–4455 https://doi.org/10.1093/bioinformatics/btz305.
GeneRax: Morel et al (2020) MBE https://doi.org/10.1093/molbev/msaa141.
PastML: Ishikawa et al (2019) MBE 36(9):2069–2085 https://doi.org/10.1093/molbev/msz131.
OpenTree: Mctavish J et al (2021) Syst Biol 70(6): 1295–1301. https://doi.org/10.1093/sysbio/syab033.
API and data structures
Topiary can also be used as an API to organize general phylogenetic workflows. It uses pandas dataframes to manage phylogenetic data, allowing it to readily connect to other data science pipelines. Further, topiary provides programmatic access to RAxML-NG, GeneRax, Muscle5, and BLAST (local and remote). It also wraps portions of the OpenTree and PastML Python APIs for convenient interaction with topiary pandas dataframes.
You can see examples of the topiary API in action inside Jupyter notebooks in the topiary-examples github repo. For a detailed description of the data structures and API, see the Data Structures and API pages.