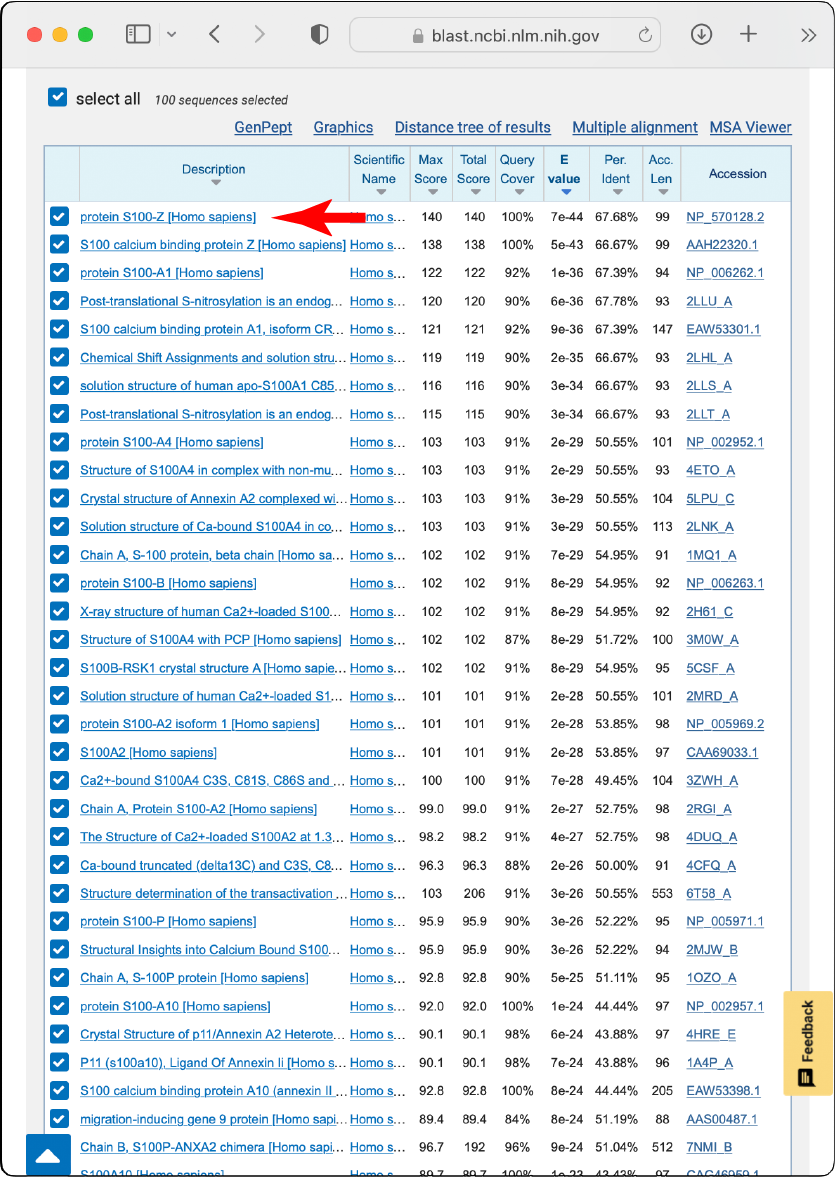
Determining taxonomic scope
The following steps can be used to determine the taxonomic scope for a protein of interest. We recommend doing this for non-microbial proteins. For microbial proteins, the species tree is poorly defined; therefore, topiary automatically sets the scope to “all bacteria” or “all archaea”.
BLAST known protein sequences against the non-redundant clustered BLAST database, setting the
Max target sequencesparameter to 1,000 or more.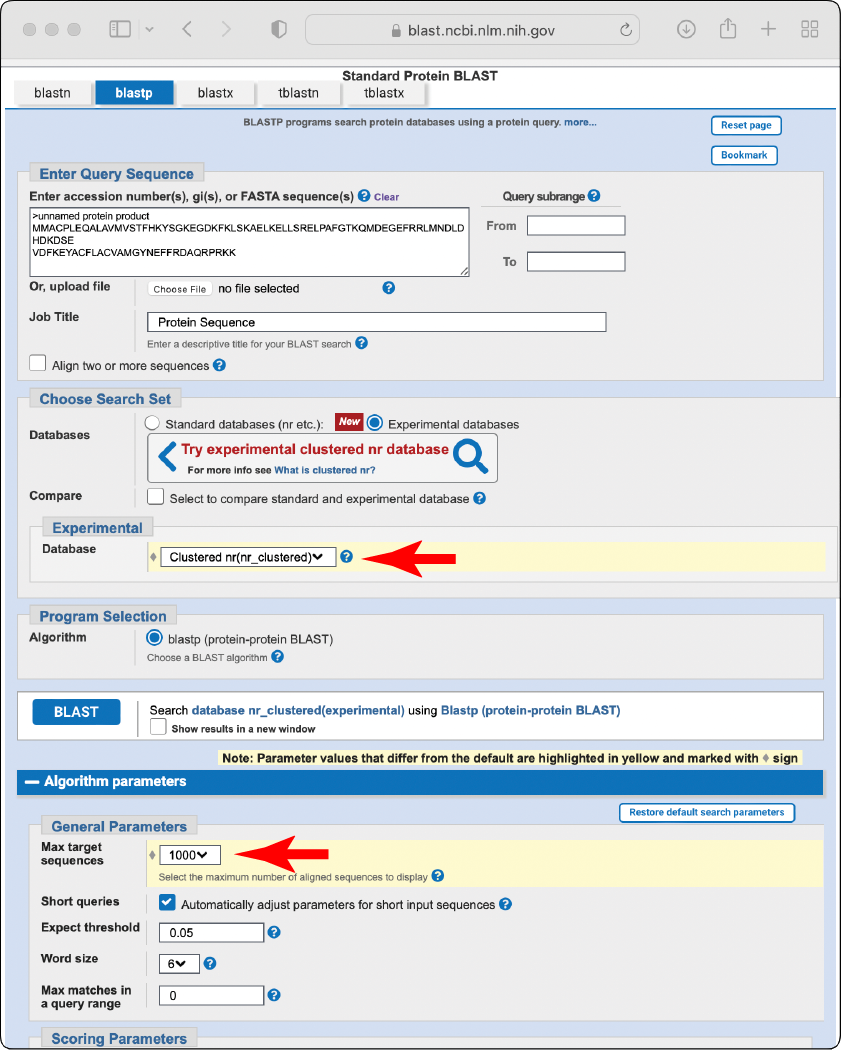
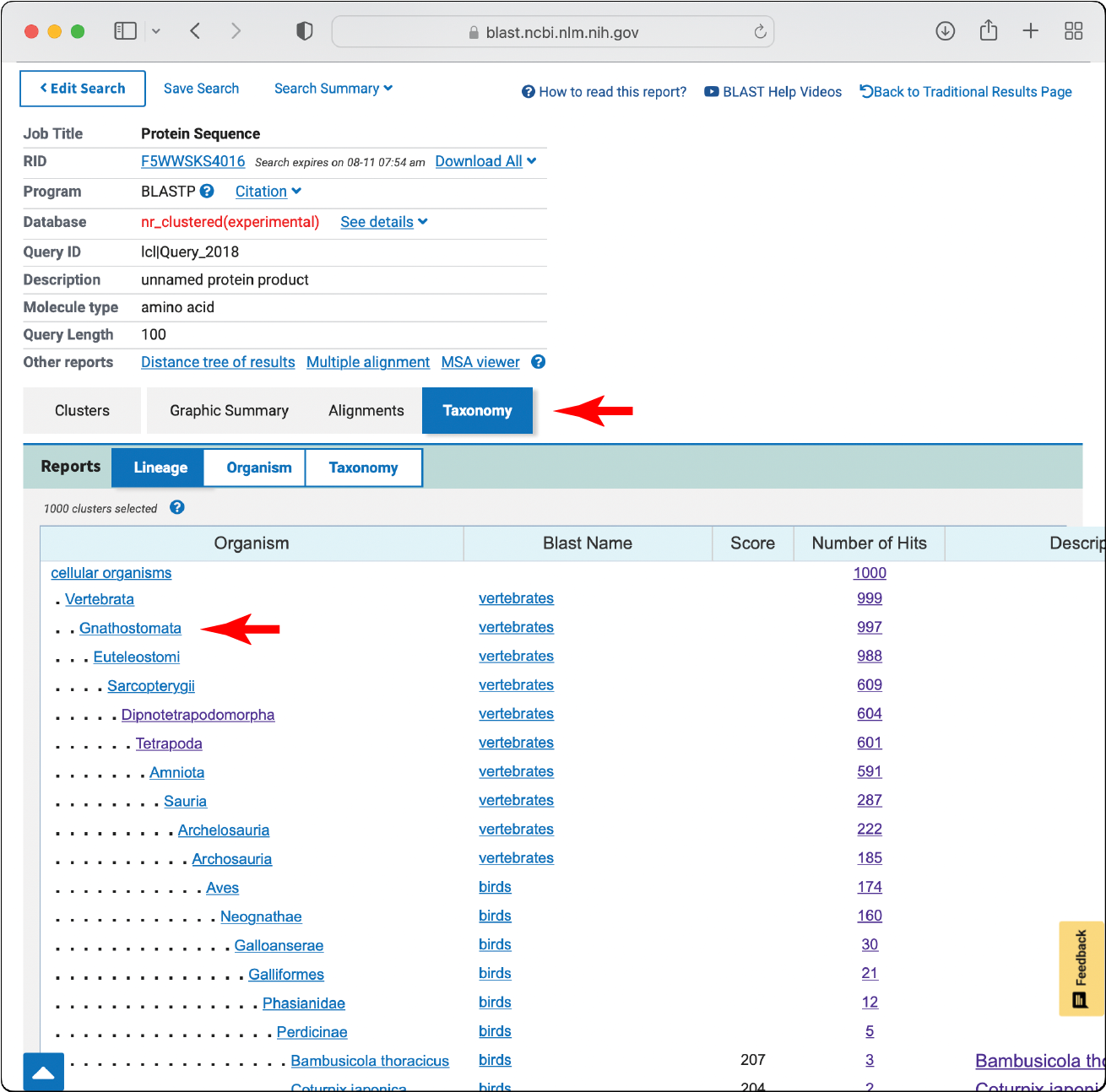
Take a few representative sequences from the most divergent species in the outputs. These can be selected using the “Taxonomy” tab.
BLAST the divergent sequences back against the NCBI non-redundant database, limiting the search to the species from which you took your known sequences. (If, for example, you used a human sequence as your starting point, you would limit this “reciprocal” query to Homo sapiens.)
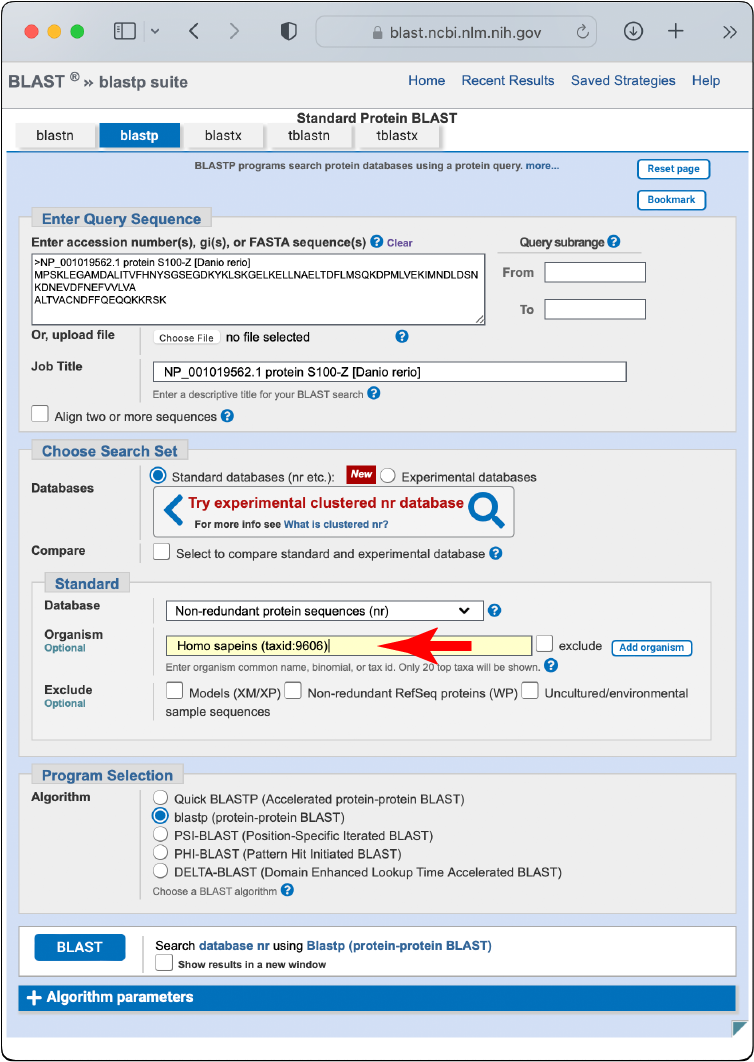
If this BLAST search pulls up your starting protein as a top hit, it is good evidence that the species from which the sequence came has the protein and should be included in the taxonomic scope.